Chapter 6 Clustering
The default option is to arrange the rows and columns in the same order as they are provided. We saw in a previous section that it is easy to arrange the rows and columns into a clustered formation using the pretty.order.rows and pretty.order.cols arguments.
6.1 Dendrogram
It is natural to supply a dendrogram that highlights the hierarchical clustering of the columns and/or rows using the col.dendrogram and row.dendrogram arguments. Note that if you want to implement the row or column ordering implied by the dendrogram, but to remove the dendrogram itself, you can use the pretty.order.rows and pretty.order.cols arguments.
superheat(mtcars,
# scale the matrix columns
scale = TRUE,
# add row dendrogram
row.dendrogram = TRUE)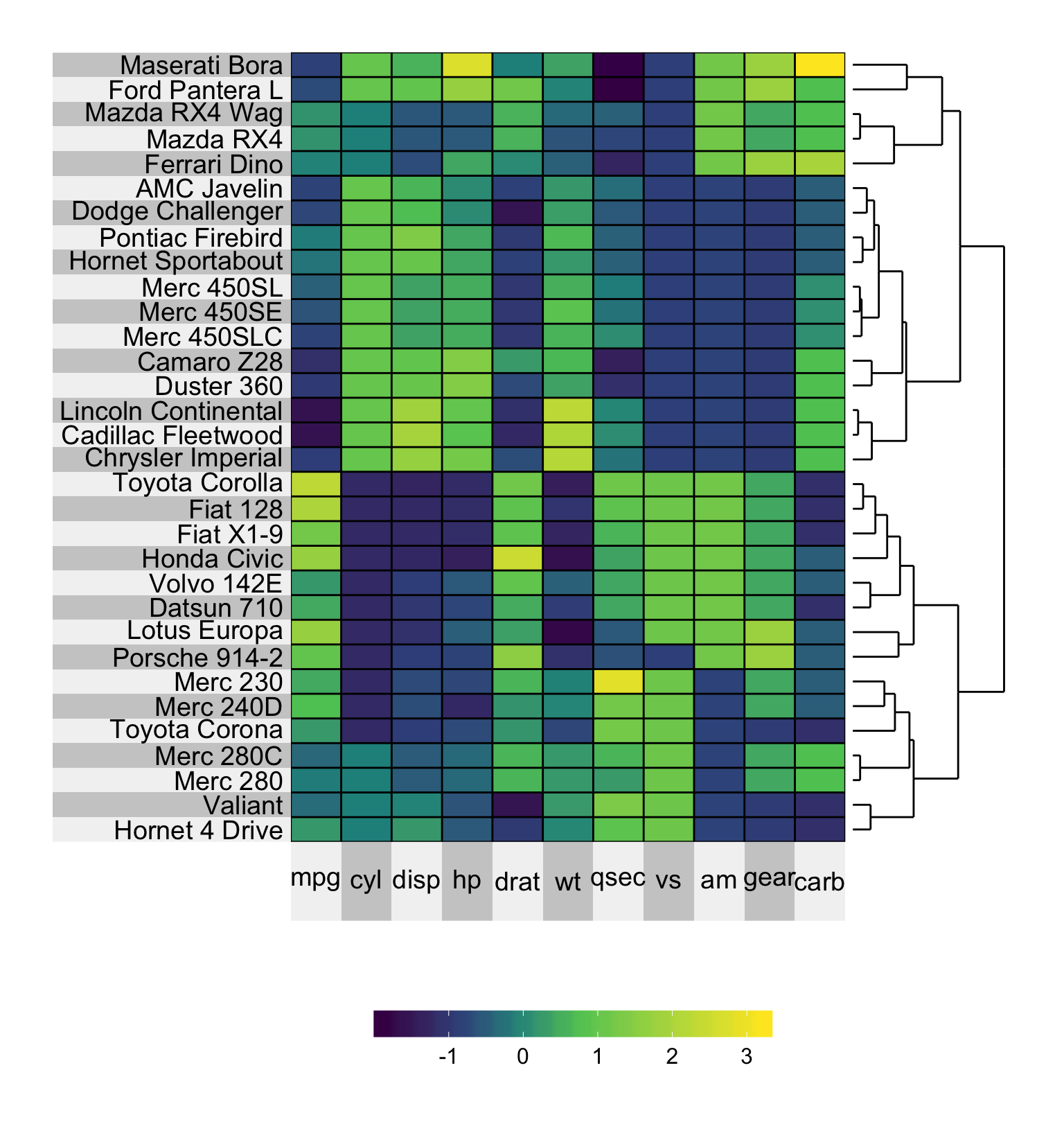
6.2 Generating clusters
Grouping the rows and/or columns into a pre-specified number of clusters is a nice way to highlight structure and simplify visualization. For example, we can group the rows into three groupings by specifying n.clusters.rows = 3. The underling clustering algorithm is kmeans(), but you can use hierarchical clustering by specifying clustering.method = 'hierarchical'.
In order to get the same clustering every time you must set the seed or provide your own clustering membership vector.
set.seed(2016113)
superheat(mtcars,
# scale the matrix columns
scale = TRUE,
# generate three column clusters
n.clusters.rows = 3)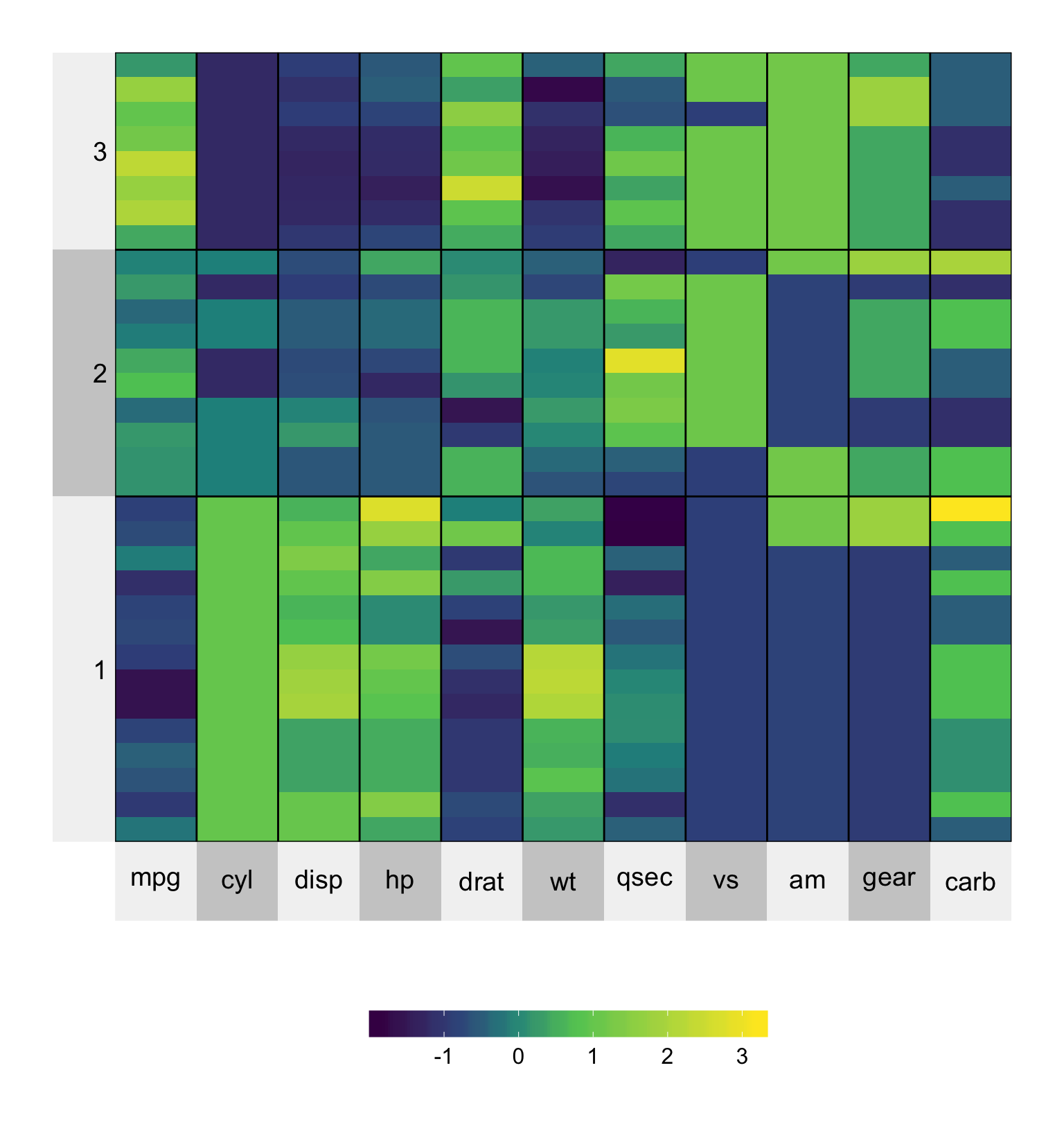
By default, when clustering the corresponding labels are grouped into the cluster name (typically 1, 2, 3, … etc). If you would like to force the labels to be the original variable names, you can specify left.label = 'variable' or bottom.label = 'variable, depending on whether it is the left or bottom labels, respectively.
set.seed(2016113)
superheat(mtcars,
# scale the matrix columns
scale = TRUE,
# generate three column clusters
n.clusters.rows = 3,
left.label = 'variable')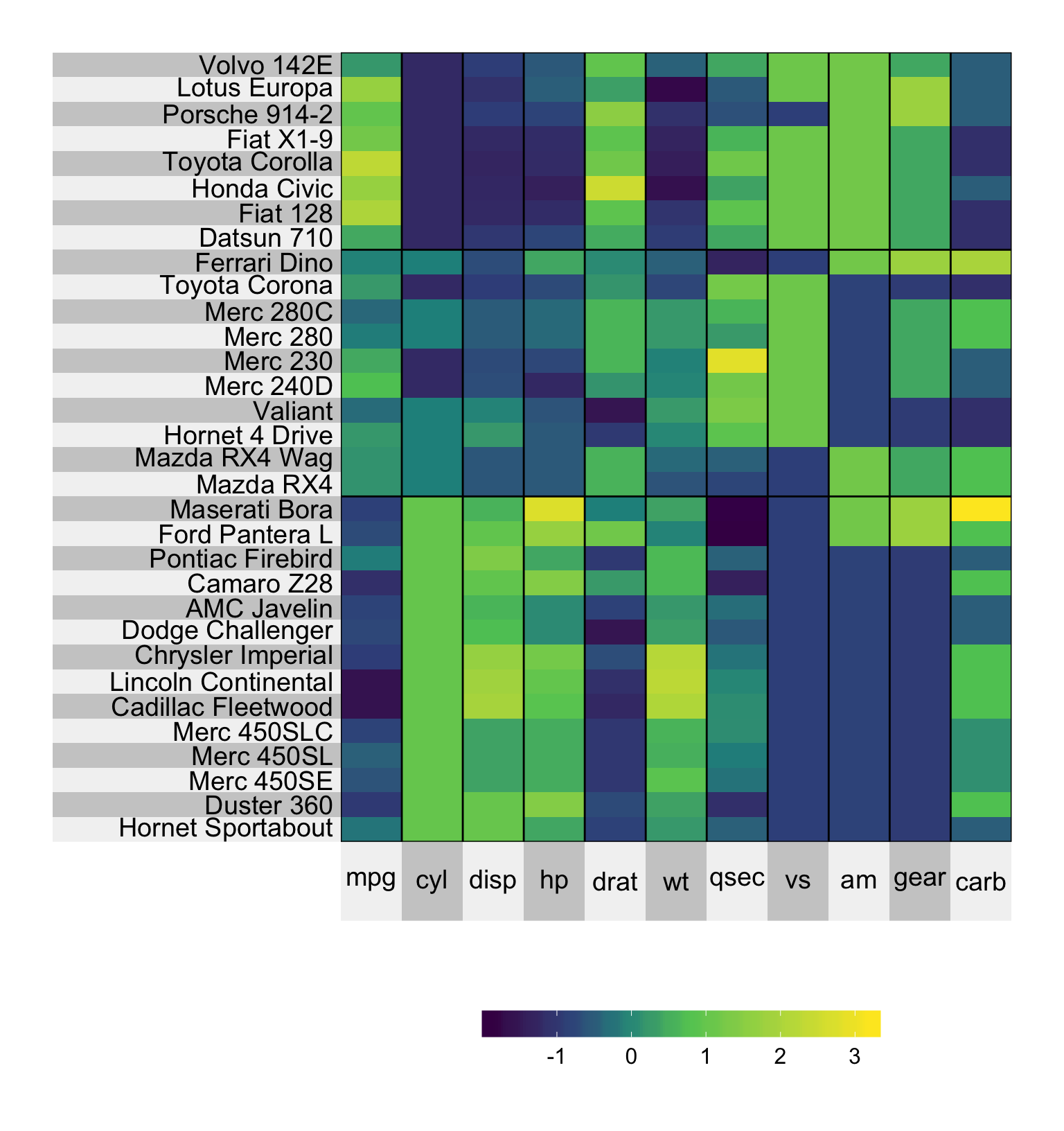
6.2.1 Extracting the clusters
If you would like to be able to extract the clusters generated by the superheat() function, then you need to first save the superheat object as a variable. From this variable, you can access the clusters that are stored in the membership.rows and membership.cols entries.
set.seed(2016113)
superheatmap <- superheat(mtcars,
# scale the matrix columns
scale = TRUE,
# generate three column clusters
n.clusters.rows = 3,
left.label = 'variable',
print.plot = F)
# extract the clusters
superheatmap$membership.rows## Hornet Sportabout Duster 360 Merc 450SE
## 1 1 1
## Merc 450SL Merc 450SLC Cadillac Fleetwood
## 1 1 1
## Lincoln Continental Chrysler Imperial Dodge Challenger
## 1 1 1
## AMC Javelin Camaro Z28 Pontiac Firebird
## 1 1 1
## Ford Pantera L Maserati Bora Mazda RX4
## 1 1 2
## Mazda RX4 Wag Hornet 4 Drive Valiant
## 2 2 2
## Merc 240D Merc 230 Merc 280
## 2 2 2
## Merc 280C Toyota Corona Ferrari Dino
## 2 2 2
## Datsun 710 Fiat 128 Honda Civic
## 3 3 3
## Toyota Corolla Fiat X1-9 Porsche 914-2
## 3 3 3
## Lotus Europa Volvo 142E
## 3 36.3 User-supplied clusters
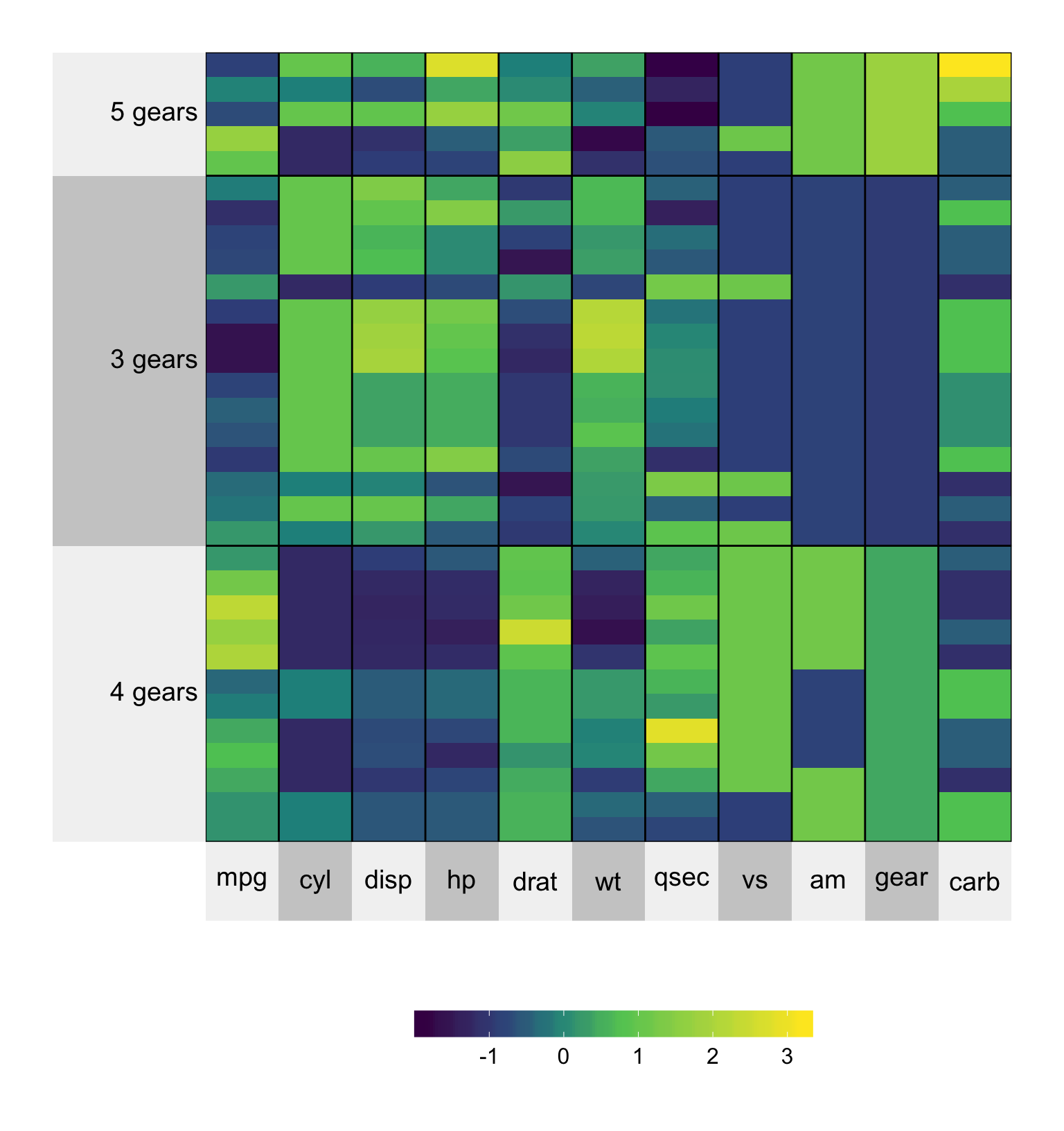
The best way to conduct clustering on your matrix is to provide a pre-specified membership vector using the membership.rows/membership.cols argument. Suppose, for our mtcars example, we wanted to group by number of gears.
gears <- paste(mtcars$gear, "gears")
set.seed(2016113)
superheat(mtcars,
# scale the matrix columns
scale = TRUE,
# cluster by gears
membership.rows = gears)